|
19. DNA Replication
|
Tweet |
|
|
 |
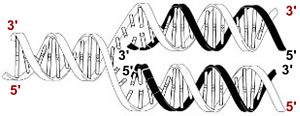
The double-helical structure of DNA immediately suggested to Watson and Crick a simple mechanism for the accurate duplication of the genetic information stored in DNA. Each strand contains all of the information necessary to specify the sequence of its complementary strand. |
|
Polymerization (always) involves the reaction of an incoming deoxynucleotide triphosphate (dNTP) with the 3' hydroxy group of the pre-existing polynucleotide chain [link]. The reaction releases pyrophosphate (P-P) and forms a phosphodiester bond. The 3' OH of the added dNTP becomes the new 3' OH of the growing polynucleotide. |
 |
|
The sequence of a polynucleotide is always written in the 5' to 3' direction, just as the sequence of a polypeptide is always written from its N-terminus to its C-terminus (as we will see).
Both DNA and RNA polymerases synthesize nucleic acids by adding nucleotides to the 3'-OH end of a molecule. |
|
DNA replication is a complex process - the steps are mediated by various proteins. In the bacterium Escherichia coli over 100 genes are involved in DNA replication and repair. Replication begins at specific sequences along the DNA strand, known as origins of replication or origins for short. Origin sequences are recognized by specific proteins. The binding of these proteins initiates the assembly of an origin recognition complex, an ORC. In the laboratory, DNA is generally heated to 94-100°C in order to insure complete denaturation (strand separation). In the cell, various proteins act on the DNA to locally denature (unwind) the DNA to form a replication bubble. A multiprotein complex will assemble at each end of the replication bubble, these structures are known as replication forks. |
 |
Using a single replication origin and two replication forks moving in opposite directions, a growing E. coli can replicate its ~4,700,000 base pairs of DNA in ~40 minutes. |
|
Each replication fork moves along the DNA synthesizing ~1000 base pairs of DNA per second Synthesis is an accurate process; the polymerase makes about one error for every 10,000 base pairs it synthesizes. Most of these errors are quickly recognized, since they lead to the formation of a base mismatch. When a mismatched base pair is formed, the DNA polymerase recognizes its presence, reverses and removes it (using an exonuclease activity), and than resynthesizes it, correctly. This process is known as proof-reading; the proof-reading activity of the DNA polymerase complex reduces the total DNA synthesis error rate to ~1 error per 1,000,000,000 (109) base pairs synthesized. |
|
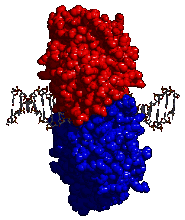
DNA replication and molecular machines: The process of nucleic acid replication involves a number of molecular machines, we will consider only one as an example - the clamp loader. Once DNA replication begins it is important that the polymerase complex remains attached to the DNA. This is accomplished by the assembly of a doughnut shaped protein, known as a sliding clamp, around the DNA double helix. |
 |
 |
The clamp is added to DNA by a protein known as a clamp loader [link][link]. Once closed around the DNA the clamp diffuses freely along the length of the DNA molecule, but it cannot leave the DNA. The rest of the polymerase complex is attached to the clamp. This keeps the polymerase complex from drifting away from the DNA. The attached clamp-polymerase complex now assembles many nucleotides into a new DNA strand before it falls off. |
| DNA-dependent, DNA polymerases cannot start a new DNA chain de novo or from nothing. They can only add nucleotides to a pre-existing DNA (or RNA) strand. Why this is, evolutionarily, is not known, but it is a conserved (homologous) feature of the DNA polymerases involved in DNA replication. |
|
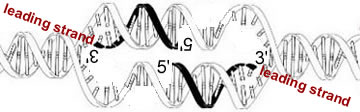
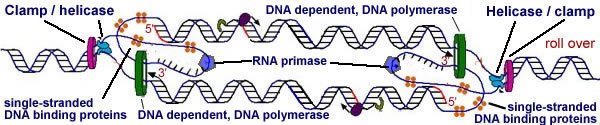
When the DNA helix opens there are no pre-existing DNA strands for the DNA polymerase to extend. So, how does synthesis start? The answer is that a short RNA molecules, known as a primers, complementary to the two strands of the open DNA, are synthesized by a DNA-dependent, RNA polymerase - this enzyme is known as primase. |
  |
|
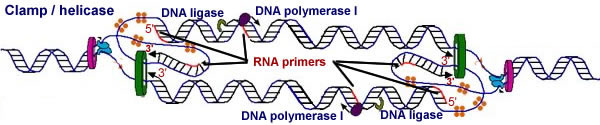
DNA polymerase synthesizes DNA onto the ends of these primers in the standard 5' to 3' manner. These first primers define the leading strands of DNA synthesis. As the DNA forks move, more DNA is unwound, and new primers have to be synthesized to insure replication of the "lagging strands"(go to this video: quite nice,although very "flat"). No new primers have to be synthesized on the leading strand. Later in the DNA replication process, the RNA primers are digested away, replaced with DNA, and joined together by the enzyme DNA ligase to produce a single, uninterrupted replicated strand. |
 |
|
Eukaryotic cells can contain more than 1000 times the DNA found in a typical bacterial cell. Moreover, the eukaryotic DNA replication enzyme complex is much slower (about 1/20th as fast) than the prokaryotic system. While a bacterial cell can replication is circular, ~3 x 106 base pair chromosome in about 1500 seconds using a single origin of replication To replication their DNA eukaryotic cells use multiple origins of replication, scattered along the length of each chromosome. |
Another topological complexity is associated with the replication (and segregation) of closed circular DNA molecules, as well as extremely long linear DNA molecules. The replication of DNA unwinds the DNA, and this unwinding requires topoisomerases to allow replication to continue. |
| |
A specific molecular machine acts when replication forks "crash" into one another. It resolves the strands and links them, leading to a single double stranded DNA molecule per chromosome. So even though they may contain 1000 times more DNA than E. coli, DNA replication can still be completed in as little as 10 minutes. ← A video of the complex process of DNA replication (something to look forward to in Molecular Biology!) |
|
|
|
Questions to answer
|
|
|
Questions to ponder
|
|
|
|
|
|
| replace with revised beSocratic activity |
|
|
|
modified
10-May-2014
|