| 25. Genomes & genes |
|
|
The larger the genome, the more information it can, at least in theory, contain. The organism with the largest known genome is the plant, Paris japonica (Pellicer et al, 2010), with an estimated size of ~150,000 x 106 (millions of) base pairs, which compares to the ~3,200 x 106 base pairs of humans (which encodes approximately 25,000 genes). |
Smaller genomes are found in bacteria and archaea, but the smallest genomes occur in obligate parasites and endosymbionts. For example, the bacterium Mycoplasma genitalium, the cause of non-gonococcal urethritis, contains ~0.580 x 106 base pairs of DNA, which encodes ~500 distinct genes. It has the smallest genome found for an organism that can be grown independently. Using mutagenesis studies, Lewis et al (2006) found that 382 of its genes are essential, of which ~28% had unknown functions. The obligate endosymbiont, Carsonella ruddii has 159,662 (~0.16 x 106) base pairs of DNA encoding "182 ORFs (open reading frames or genes), 164 (90%) overlap with at least one of the two adjacent ORFs" (Nakabachi et al, 2006). |
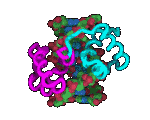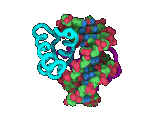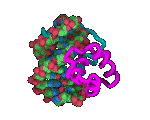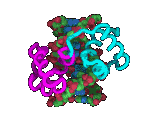
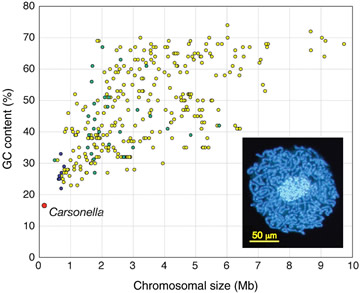 Relationship
between genome sizes and GC content of 358 complete genomes from Bacteria
and Archaea. In the insert, the tubular structures are Carsonella living
inside a cell of a phloem sap-feeding insect Pachypsylla venusta -
the host cell's nucleus is the larger blue oval. (Nakabachi
et al., 2006) Relationship
between genome sizes and GC content of 358 complete genomes from Bacteria
and Archaea. In the insert, the tubular structures are Carsonella living
inside a cell of a phloem sap-feeding insect Pachypsylla venusta -
the host cell's nucleus is the larger blue oval. (Nakabachi
et al., 2006) |
|
Locating information within DNA: The DNA molecules that contain genes can be thousands to millions of base pairs in length. More to the point, a single DNA molecule can contain hundreds to thousands of genes. To access the information stored in a gene, it is necessary to find the sequence of the gene so that it can be transcribed into RNA. This sequence has to be recognized and distinguished from all of the other DNA sequences present within the cell. This is accomplished by using a two-component system. The first part of this system are specific nucleotide sequences within the DNA (very much like the origin of replication sequences we discussed before.) |
These sequences provide a molecular address that can be used to identify the specific region of a specific strand of the DNA to be transcribed. The set of sequences involved in identifying the region of a gene to be transcribed is known as the gene's regulatory region. The regulatory region of a gene can be simple and relatively short or complex and extremely long. In some human genes, the gene's regulatory region is spread over thousands of base-pairs of DNA, located both "up-stream" and "down-stream" of the coding region. You can carry out a simple mathematical exercise, how long would a nucleotide sequence have to be, to be unique within 3,200,000,000 base pairs? |
 |
Regulatory DNA sequences are recognized by proteins that bind to DNA in a sequence specific manner; these proteins are known as transcription factors because they regulate transcription. Specific regulatory sequences are bound by specific transcription factors, and it is common that multiple transcription factors are used together (combinatorially) to recognize and regulate the expression of specific genes. |
A specific transcription factor can act either positively or negatively. Positively-acting transcription factors, known as activators, stimulate transcription. Negatively-acting transcription factors, known as repressors, suppress transcription. Sometimes a transcription factor that acts positively at one DNA regulatory site will act negatively at another, depending upon its interactions with other proteins that bind near by, or that associate with the transcription factor. Transcription factors are ""context dependent". |
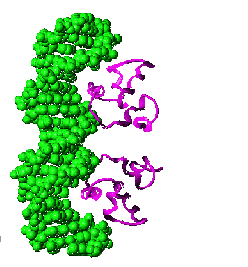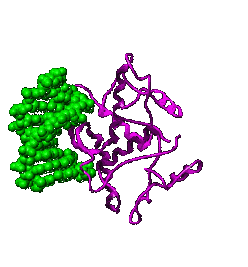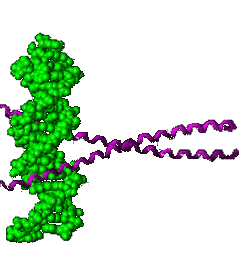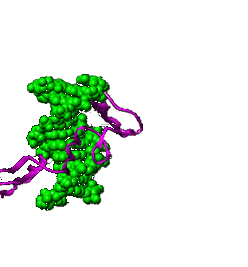 |
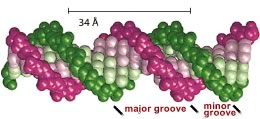
DNA-binding transcription factors recognize specific DNA sequences by interacting with the surfaces of the base pairs visible in the major and minor grooves of the DNA helix. A single base
pair change in a gene's regulatory region can profoundly alter the
efficiency or timing of transcription. |
|
 |
The binding of transcription factors recruits or inhibits the recruitment of an enzyme complex, the DNA-dependent, RNA polymerase, to the DNA. This RNA polymerase synthesizes RNAs. |
Where RNA polymerase starts transcribing RNA is known as the transcription start site. Where it falls off the DNA, and so stops transcribing RNA, is known as the transcription termination site. As transcription initiates, the RNA polymerase moves away from the transcription start site. Once the RNA polymerase complex moves far enough away (clears the start site), there is room for a new polymerase complex to associate with the DNA, through interactions with transcription factors. The efficiency of RNA polymerase binding and activation determines the rate at which RNA molecules are produced. |
Questions to answer
|
|
Questions to ponder |
|
| replace with revised beSocratic activity |
Tweet
revised
10-May-2014
|