| 26. Regulatory networks |
Inside the cell, DNA exists in association with various proteins. This DNA-protein complex is known as chromatin. Based on the structure of DNA, each base pair is about 0.34 nm in length. A kilobase (that is, 103 base pairs) of DNA is therefore about 0.34 µm in length. A bacterium, like E. coli, has ~ 3 x 106 base pairs of DNA – that is a DNA molecule almost a millimeter in length. |
 |
The bacterial
cell itself
is only about 0.8 µm in diameter and about 2 µm
in length. A
human cell has about 6000 times more DNA, that is a total
length of greater than 2 meters. To fit
the DNA molecule into a cell takes some packing. The
way the DNA is assembled into chromatin, particularly in
eukaryotic cells, can dramatically influence the ability
of transcription factors to interact with and bind to regulatory
sequences. |
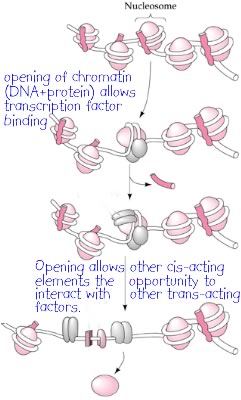 |
Regions of chromatin that are packaged so as to be inaccessible to regulatory proteins are known as heterochromatin. Chromatin that is accessible and transcriptionally active is known as euchromatin. Some regions of chromatin are folded away more or less permanently; they are unfolded only during DNA replication. These are known as constitutive heterochromatin. Other regions can be unpacked in different cell types and are known as facultative heterochromatin. A particularly dramatic example of this process occurs in female mammals. One of the two X chromosomes is packed into a heterochromatic state, known as a Barr body. Most of the genes on this heterochromatic X chromosome are not expressed; so even though females (XX) have twice as many of the genes located on the X chromosome as males (XY), both make about the same amount of gene product. |
Transcription factors also influence the proteins that act on chromatin to make the DNA within it accessible or inaccessible. This regulation of chromatin packing can act on blocks of DNA, regulating all of the genes within the affected region. In the end, the gene regulatory elements within the DNA compete with one another for the binding of DNA-dependent, RNA polymerases. Genes that assemble the most efficient promoters are the most effective at loading RNA polymerase molecules. These genes will produce more copies of the RNA that they encode. |
Networks: A cell's behavior will be determined in large part by which of its genes are actively expressed. How can a cell determine that the "right" set of genes is activated for a particular situation? Clearly feed back systems are needed. Genes that need to be expressed together to perform a function can be co-regulated by using similar sets of regulatory sequences. |
 |
| A comprehensive analysis of the interactions between 106 transcription factors and regulatory sequences in the baker's yeast Saccharomyces cerevisiae revealed the presence of a number of regulatory motifs. |
| Autoregulatory loops: In this type of regulatory motif, a transcription factor binds to regulatory sequences that regulate its own transcription. Such interactions can be positive (amplifying) or negative (squelching). | 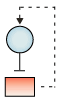 |
|
| Feed forward interactions: A transcription factor regulates the expression of a second transcription factor; the two transcription factors then cooperate to regulate the expression of a third gene. | 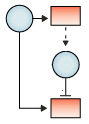 |
|
← Single and multiple input modules: A transcription factor binds to sequences in a number of genes, regulating their coordinated expression. In most cases, sets of target genes are regulated by sets of transcription factors that bind in concert. Regulation of regulatory networks:→ Transcription factors are proteins, and so their stability and activities can be regulated by interactions with other proteins, allosteric factors, and post-translational modifications. It is through such interactions that signals from outside the cell can alter the patterns of gene expression. |
  |
At the same time, internal signals, generated by metabolic processes, can also feedback to the gene regulatory system in order to maintain the homeostatic state. In fact, such feedback is critical to maintaining regulatory networks – inhibiting them from spiraling out of control – which would lead to death. |
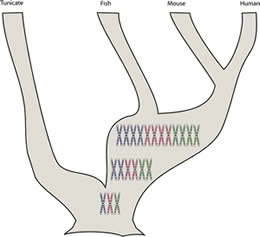
Genome and gene duplication: An obvious question is, how can biological systems increase their complexity? Where do new genes and new activities come from? One mechanism is by duplicating pre-existing genes [link]. Typically selection will act to maintain the function of one copy of the gene (which was important in the organism before the duplication and remains so afterward). The second copy is then freer to change. Mutations may lead to the loss of a functional gene product, or a gene product with altered properties. |
 |
There a number of different types of duplication events, which range from the duplication of a small region of a specific chromosome to the duplication of the entire genome (we will not go into the molecular details involved). The advent of genomic sequencing and the comparison of genomes from different organisms suggests that many types of organisms have undergone both processes. The most dramatic type of duplication event is the duplication of an entire genome. After such an event (which is rare), some genes will be lost. This will be the result of random mutations that inactivate the gene. Since there is another copy of the gene, the loss of the second copy may have little effect. On the other hand, when a gene is duplicated, it is likely that the total amount of gene product produced will increase - in some cases, increased gene expression can have harmful effects (think mammalian X chromosome and females). When this is the case, inactivation of one copy of the gene may, in fact, be beneficial. Mutations can also lead to changes in activity and specificity - a mutation can change a protein so that it can bind new substrates and catalyze new reactions, a process known as neofunctionalization. One issue with "blind" duplication events is that the
development of new functions occurs only after duplication. The
duplicated gene may be lost through mutation (or genetic drift).
As we look at the genes of various organisms, we find evidence for both gene and genome duplication. For example, compared to their invertebrate ancestors, the vertebrate lineage appears to have gone through two rounds of whole genome duplication. |
Questions to answer
|
|
Questions to ponder |
|
| replace with revised beSocratic activity |
Tweet
revised
10-May-2014
|