| 27. Cell growth & division |
|
|
Perhaps the most characteristic feature of life is the ability to replicate, to make copies of itself. During the process of cell division, the genetic material must be replicated. The two strands of the DNA molecule separate locally, and each serves as a template for generating a new strand. |
Changes in the DNA that accumulate prior to, or which occur during replication, are passed on to the daughter cells. These daughter cells are built using energy and matter captured from the environment. |
If we inoculate a culture with a few bacteria, within a few hours they will have transformed components of the medium into millions of copies of themselves. When we look at these cells with a microscope, we find that they do not grow haphazardly. They appear remarkably uniform in both size and shape. Different types of cells have characteristic shapes and sizes. They grow to a certain size and then divide. |
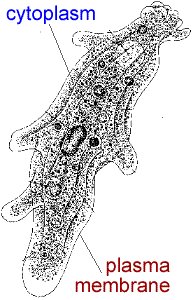
Cells monitor and control their size. For example, consider the single celled eukaryote, Amoeba proteus – these organisms divide only after they have grown to a characteristic size. Using microsurgical methods, it is possible to keep cells small by simply repeatedly cutting off parts of the cell. After the cell heals, it grows. If you do this repeatedly, the cell grows but never divides. This type of experiment argues that the cell does not use time to decide to divide, but rather size. [These studies were carried out by David Prescott, one of the founders of MCDB; he also used the fact that one can experimentally remove the nucleus of these cells to show that RNA was made in the nucleus!] |
 |
How does the cell know when it has reached the correct size, how does it know when to divide? Studies on yeasts (unicellular eukaryotes) have identified mutations that lead to cells that are consistently larger or smaller than the usual, or wild type phenotype. The ability to find such mutations implies that an active, genetically encoded system controls when cells divide. |
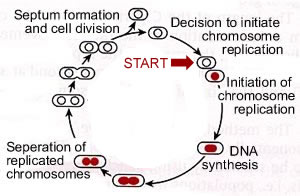
Bacterial cell cycles: The replication of a cell, or an organism, can be thought of as a cycle. Somewhat arbitrarily, we place the beginning of this cycle with the decision to replicate the genetic material, the DNA. This is a critical decision for the cell. DNA replication involves the unwinding of the DNA and a set of highly interdependent and coordinated processes. Consider the following catastrophic scenario. A cell begins replicating its DNA, but before it completes the process it runs out of resources -- ATP levels fall and the other deoxyribonucleotide triphosphates needed to synthesize DNA are in short supply. Under these conditions replication forks will stop and the DNA will be left unwound and incompletely replicated. This is a situation that is likely to lead to DNA damage. |
To avoid this possibility, cells tightly regulate the initiation of DNA replication. This decision point is known as "start"; once made, the cell begins on the path to DNA replication. In bacteria start involves the molecular decision to build a replication initiation complex. |
 |
|
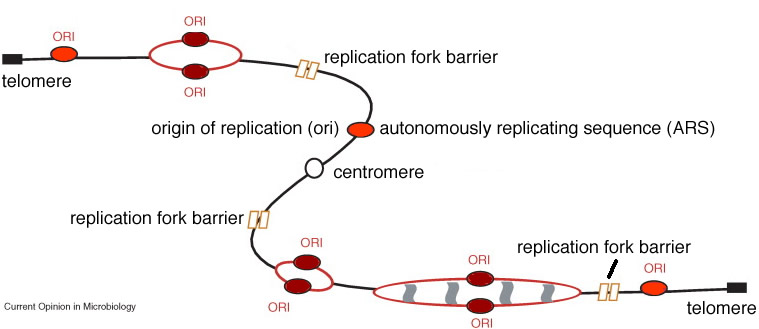
The initiation complex consists of proteins that associate with a specific DNA sequence, known as the origin of replication or origin. There is a single origin in the bacterial chromosome and it acts like a gene. In E. coli this gene is called OriC. For a DNA molecule to replicate, it must have at least one functional origin of replication. In most prokaryotes, the chromosome is a circular DNA molecule. The origin (OriC) is where the replication bubble initiates and it is associated with the plasma membrane. |
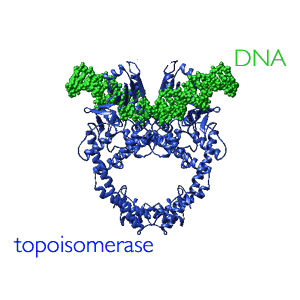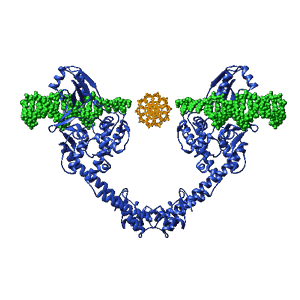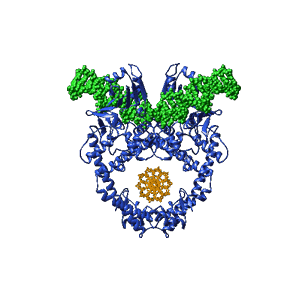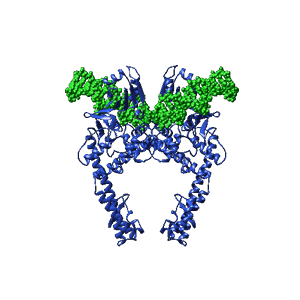
The two replication forks move away from one another and around the chromosome. The forks collide in the region of the chromosome known as the terminator region or "ter". At the end of replication, two sister chromosomes are entangled with one another, they are concatenated. They are untangled by the enzyme, DNA topoisomerase II, which can pass one double-stranded DNA molecule through another. |
 |
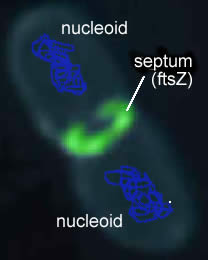 |
Once the replicated chromosomes are separated, the cell can be divided into two. A septum assembles at the center of the cell. This molecular "purse string" is linked to the inner surface of the plasma membrane. As it contracts, it pulls the plasma membrane, which then pinches off to separate the two cells. When growing under optimal conditions bacteria can divide as fast as once every 15 to 20 minutes. |
Eukaryotic chromosomes: As we move on to consider chromosome replication, segregation (mitosis) and cell division (cytokinesis) in eukaryotes, it is important to consider the differences in chromosome structure between eukaryotes and prokaryotes (bacteria and archaea). While prokaryotic chromosomes are typically circular molecules, with a single chromosome per cell, eukaryotic chromosomes's are linear, and there are typically many different chromosomes.. If you remember back to the mechanism of DNA replication (which is similar in both prokaryotes and eukaryotes), this raises issues associated with complete replication of the chromosome ends. This problem is solved by the presence of special DNA sequences (telomeres) and a specific enzyme, telomerase. There is a telomere at each end of eukaryotic chromosome. |
 |
During mitosis (see below) the centromere complex interacts with the molecular machinery that segregates the replicated chromosomes, and the connection between the replicated chromosomes is severed during mitosis. In meiosis , the centromere connection between replicated chromosomes occurs during the second meiotic division (see next reading). |
The eukaryotic cell cycle: The most dramatic event in the eukaryotic cell cycle, the one that caught the eye of early microscopists, is the drastic change in nuclear organization associated with cell division. This process of chromosome segregation is known as mitosis. As the cell enters mitosis, chromosomes appear as distinct bodies. While most prokaryotic cells have a single circular chromosome, most eukaryotes have multiple linear chromosomes, and each chromosome has multiple origins of replication. Each chromosome is a single DNA molecule. The DNA in each chromosome must be completely replicated and the copies segregated so that each daughter cell receives one and only one complete copy of each chromosomal DNA molecule |
|
The complexity of the eukaryotic cell, makes mitosis and cell division (known as cytokinesis, more mechanically intricate than the analogous processes in prokaryotes. As in the case of bacterial cells (above), eukaryotes have a cell cycle. Start is located in the period known as the G1 phase of the cell cycle; the period during which DNA synthesis occurs is known as S phase. The period between the end of S and the beginning of mitosis is known as the G2 phase of the cell cycle. Mitosis itself is known as M phase. |
|
The length of the cell cycle can vary tremendously, from hours to years. Cells that are not actively dividing are said to be in Go. Some cells enter Go and never divide again – these cells are said to be terminally differentiated. We will leave the mechanical and molecular details of mitosis and cytokinesis to more advanced classes in cell biology; what is critical to understand here is that they provide a complete copy of the genome to each daughter cell (mitosis) and they divide the cell into two (cytokinesis). |
| Checkpoints: The process of chromosome replication and segregation is so critical to the future of the replicating cell that its accuracy is checked in a number of ways. There is a DNA damage checkpoint that inhibits DNA replication until damaged DNA is repaired. The process of chromosome replication, segregation and cell division is more complex in eukaryotes, and so there are more possibilities for error. There are correspondingly more checkpoints. These include
|
The DNA replication and repair checkpoints are located throughout interphase (G1, S and G2) while the checkpoints associated with chromosome segregation are located in M phase. A cell that fails to correct errors generally undergoes a process of "programmed cell death" or apoptosis. This is a type of "fail-safe" that keeps abnormal cells from surviving. While we will not consider apoptosis further, it is a critical component of the social life of cells. Where apoptosis fails, cancer (and other hyperproliferative diseases) can occur. |
Questions to answer
|
|
Questions to ponder |
|
| replace with revised beSocratic activity |
Tweet
revised
10-May-2014
|